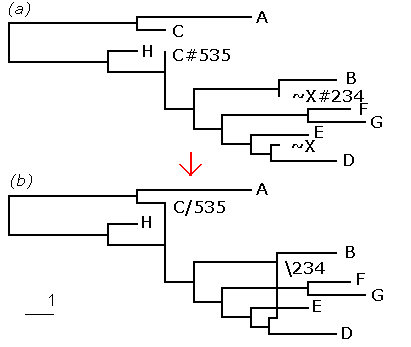
Fig. 1. The (a) tree representation and the (b) network representation of a set of recombinant sequences is shown
Serial NetEvolve 1.0: A flexible utility for generating serially-sampled sequences along a tree or recombinant network |
Patricia
Buendia and Giri Narasimhan
Bioinformatics Research Group (BioRG)
Florida International University
Serial NetEvolve is a modification of the Treevolve program in which serially sampled sequences are evolved along a randomly generated coalescent tree or network (Grassly et al. 1999; Hudson 1983; Kingman 1982) . Treevolve offers a variety of evolutionary model and population parameters including a rate of recombination and as such it was chosen over other programs to be adapted for the simulation of serially sampled data. The new features include the choice of either a clock-like model of evolution or a variable rate of evolution, simulation of serial samples and the output of the randomly generated tree or network in Newick format or in our newly formulated NeTwick format.
Here we will only list the features that differentiate Serial NetEvolve from the original Treevolve.
For information on the parameters integral to the original Treevolve v1.3, please consult with its manual that can be found here:
http://www.cecalc.ula.ve/BIOINFO/servicios/herr2/TREEVOLVE/manual.html
NetEvolve’s user interface programs loads the default parameter file or a parameter file selected by the user. After making changes to the parameters, the changes may be saved to the same or new parameter file.
The global parameters appear in the front tab control containing the settings for serially sampled data. The local parameters describing the "periods of population dynamics" appear in the grid control, where each row represents the parameters for one period of population dynamics. The command line program treevolve.exe will be launched when clicking on the button “Run Simulations."
The output tree can be viewed after the simulation process finishes, if tree files with extension “.tre” are associated with a tree viewer program. Rod Page's “Treeview” can be downloaded for free at http://taxonomy.zoology.gla.ac.uk/rod/rod.html and will automatically associate tre-files with the program during installation. If you are using another tree viewing program and it fails to open, try the Microsoft Support Page on file extensions. When using Rod Page's program TreeView, we recommend selecting the tree style “Phylogram” under “Edit|Preferences.”
The parameter file contains all the parameter settings. The new global parameters in Serial NetEvolve as they appear in the parameter file and user interface are:
[sample
size per sampling time] z6
[sampling times] p5
[internal nodes sampling
probability] i1 [0=No-sampling, 0<i<=1 sampling, >1 all]
[no
clock] k
The default parameters of Serial NetEvolve can be found in the parameter file distributed with the program or here.
This setting specifies the number of sequences to sample at each time point. When these two settings are not set, the classic Treevolve version will run and use the “Sample Size” to return sequences from the zero-time baseline.
The number of sampling times.
The probability with which sequences from the internal nodes are sampled are discussed in (Drummond and Rodrigo 2000) . If the probability is set to 0, then only sequences from the leaves are sampled. If 1 then sequences at internal nodes have the same probability as the sequences at the leaves to be randomly sampled. If larger than 1 then all internal sequences are included in the output. Any value in between 0 and 1 may be chosen as the probability and it is suggested to pick smaller probabilities with increasing sequence lengths. The default is 1 for a sequence length of 1000.
Treevolve assumes a molecular clock by default. In NetEvolve the default is “No clock” which simulates variable rates of evolution for serial samples. The no-clock option has no effect when NetEvolve is run with the original Treevolve settings (no serial sampling).
In Serial NetEvolve, the randomly generated tree or network is written to a file. When the recombination rate is zero, a tree in Newick format is written to a file. When the rate is larger than zero, a network is written to file. The recombination rate should not be set to a number higher than or too close to the mutation rate, as it will cause the Treevolve algorithm to go into an infinite loop of coalescence and recombination events.
The Serial NetEvolve tree or network is equivalent to a standard phylogenetic tree in which the internal nodes represent direct ancestors of the sequences at the leaves and the branch lengths represent the number of substitutions per site. When the clock model is assumed, branch lengths do also represent “time elapsed” since the “most common recent ancestor,” i.e. the internal node at which the branches originate. The closer (in substitutions per site and in time if the clock setting was chosen) a sequence is to the direct ancestor, the shorter the branch. Therefore, a sequence that represents (and is therefore identical to) the direct ancestor at the internal node, (an internal node sequence,) will be represented by a leaf with a branch of length zero.
The sequence IDs in the sequence alignment file and tree file contain a prefix indicating the time of sampling followed by a dot and a unique identifier, such as for example 004.2, which identifies the sequence as being from the 4th sampling time point.
In order to write a recombinant network to a file, a new format was devised, that we termed “NeTwick.”
The NeTwick Format is a variation of the Newick format, which represents trees in form of nested parenthesis (Felsenstein 1999) . This new format incorporates the additional information (breakpoint position, right/left parent) that is stored in a recombinant network while keeping things simple. Unlike tree nodes, recombinant nodes have more than one parental node. In Serial NetEvolve, we (arbitrarily) chose the left parental node of a recombinant sequence to appear twice in the NeTwick format to indicate the linking relationship. One of the copies of the left parental node appears followed by the symbol “#”, along with the breakpoint position and it represents a link, not a taxon. If the left parent was not sampled, it also appears with a “~” prefix. The advantage of making two copies of the left parental node of every recombinant node is that the network can then be represented by an equivalent tree. The tree can be viewed using any tree-viewing program; a network viewer is currently being developed.
Examples
Figure 1 shows a network with 9 taxa and its tree equivalent. The left parent X was not sampled (indicated by the ~), but is present in the tree to indicate the linking relationship as shown in the network. In the proposed network representation a backward (forward, respectively) slash followed by the breakpoint number indicates whether the left parent is below (above, respectively) in a horizontally drawn network.

Fig.
1. The (a) tree representation and the (b) network representation of a set of
recombinant sequences is shown
The network in the example of Fig 1 is written to a file in the following format:
((A:4,C:1):5,((((B:3,~X:1):2,((E:2,(~X#234:0,D:2):1):1,(F:1.5,G:2):3):1):1,C#535:0):2,H:1):4);
Fig.2 shows the tree and the proposed network representation for a network with with 3 time points (5,6,7) and 3 samples per time point. The prefix of the Sequence\Taxon Id identifies the sampling time point. The left-hand parental sequence 6.10 from time 6 was not sampled (indicated by the ~), but necessary to indicate the linking relationship as shown in the network, where the sequence does not show up.
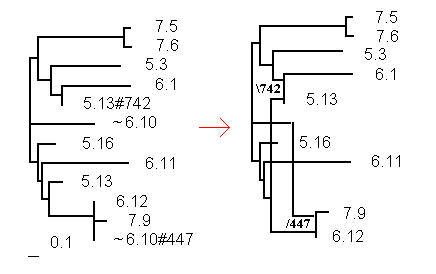
Fig. 2. Tree to network representation
for serial samples
The network in the example of Fig. 2 is written to a file in the following format:
((((6.1:0.667443,5.13#742:0):0.107317,5.3:0.674467):0.125209,(7.5:0.057291,7.6:0.062038):0.840114):0.084175,(~6.10:0.628615,((6.11:0.843106,((6.12:0,(7.9:0.112986,~6.10#447:0):0):0.439822,5.13:0.125210):0.068323):0.041674,5.16:0.167978):0.078796):0.002698);
Click here to download the Windows GUI version Serial NetEvolve 1.0
The source code of the simulator is in C. Due to limited computational and labor resources, it has only been tested on a Linux and a Windows 2000/XP computer. The C code runs in command-line mode only. I am unaware of any reason it would not work with any ANSI-compliant C++ compiler, but tweaking of code and Makefiles will likely be required. The C source code and parameter file are available here:
Click here to download the C source code of the Serial NetEvolve 1.0 simulator
Questions, comments, and bug reports may be sent to the author at pbuen001@fiu.edu. Please note, however, that development of this code is a research project, which is aimed at creating theoretical methods for phylogenetic research, not at producing production quality code. This code is being released to allow others to review, experiment with, and improve upon these methods. The code and all associated materials are provided as is, with no warranty of any kind, explicit or implicit, and no explicit or implicit promise of support.
Results of experiments with serially-sampled data generated by Serial NetEvolve can be viewed here
Many thanks to Andrew Rambaut for recommending Treevolve and making the source code available.
Drummond, A. and A. G. Rodrigo. 2000. Reconstructing genealogies of serial samples under the assumption of a molecular clock using serial-sample UPGMA (sUPGMA). Molecular Biology and Evolution 17:1807-1815.
Felsenstein, J. 1999. The Newick tree format: http://evolution.genetics.washington.edu/phylip/newicktree.html
Grassly, N., et al. 1999. Population dynamics of HIV-1 inferred from gene sequences. Genetics 151:427-438.
Hudson, R. R. 1983. Properties of a neutral allele model with intragenic recombination. Theoretical Population Biology 23:183-201.
Kingman, J. F. C. 1982. The coalescent. Stoch. Process. Appl. 13:235-248.
(Last modified: July 05, 2006 )